About me
I am a structural biologist and got attracted to machine learning after using deep learning algorithms cryoYOLO, a computer vision program to detect samples in cryoEM image, and AlphaFold2, a program to predict 3D structures of proteins or protein complexes based on their amino acid sequences. So, I attended UCSD Machine Learning Engineering Bootcamp and completed it in 2023.In the bootcamp, I developed a computer vision program named Cell Segger as my capstone project. Cell Segger uses a pre-trained UNet (a Convolutional Neural Network) model to process cell microscope images and generate cell masks binary images.Thanks for visiting my portfolio.
Projects
Structural studies on the H/ACA ribonucleoprotein complexes
Skills: X-ray crystallography | Protein purification | RNA transcription | Protein-RNA complex reconstitution | Biochemistry
The H/ACA ribonucleoprotein (RNP) complexes existing in both archaea and eukaryotes are responsible for pseudouridylations in ribosomal and spliceosomal RNAs. They are composed of four proteins CBF5, NOP10, NHP2 (in eukaryotes) / L7AE (in archaea) and GAR1, and one H/ACA RNA. Mutations in human CBF5 and NOP10 had been found linked to dyskeratosis congenita (DC), a bone marrow failure disorder. In mammals, H/ACA proteins are also the essential constituents of telomerase, a key player to maintain telomeres at the ends of chromatin. I performed structural and biochemical studies on the H/ACA RNP and had achieved the followings.
1. Determined the structure of a RNA substrate bound H/ACA RNP
2. Determined the structures of yeast H/ACA protein complexes
3. Determined the structure of H/ACA RNP chaperone Shq1
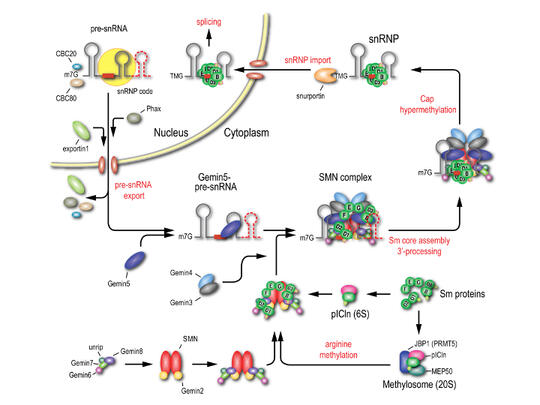
Structural studies on the human SMN complex
Skills: CryoEM | Insect cell culture | Mammalian cell culture | Comlex purification
The SMN (Survival Motor Neurons) complex, comprising SMN, Gemin2-8 and unrip, is an RNA-protein (RNP) assembly hug that is particularly well-characterized for its crucial role in the biogenesis of small nuclear ribonucleoproteins (snRNPs), key components in mRNA processing. Dysfunction of the SMN complex resulted from deletion of or point mutations in SMN accounts for an early onset motor neuron degenerative disease, spinal muscular atrophy (SMA). Lack of structural information of the SMN complex has hampered our understanding on the molecular details in the snRNP biogenesis. Therefore, I performed structural and biochemical studies and had following achievements.
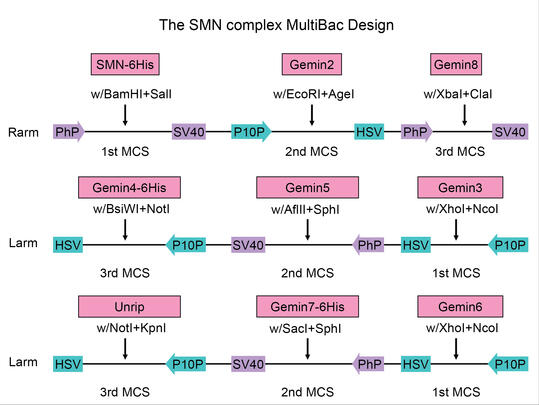
1. Constructed a Multi-Baculovirus expression system
2. Negative TEM on the purified SMN/Gemin2/6/7/8
Structural and Biochemical studies on SMN polymers
Skills: CryoEM | Single Particle Reconstruction | SEC-MALS | AlphaFold | Native PAGE
SMN forms the oligomeric centerpiece of the SMN complex. Human SMN [294 amino acids] N-terminal half contains the binding domain for Gemin2 and a Tudor domain which recognizes methylated arginines. The C-terminal half contains a three patches of poly-proline and a tyrosine-glycine-rich domain (YG), the oligomerization module, which is the most evolutionarily conserved part in SMN and accommodated for most of the known SMA-pathogenic mutations. Due to lack of structural information on SMN oligomers, it is still mysterious that how the point mutations in the YG domain affected the SMN complex functions. Therefore, I reconstituted SMN polymer, performed biochemical and structural studies on them, and achieved the followings.
1. Characterized the sizes of purified SMN polymers
2. SMN polymers are spherical nano-particles.
Reconstructed SMN oligomer from a tilt-series
3. Gemin8 stabilizes SMN polymers.
Cell Segger: A Machine Learning-based Cell Instance Segmentation Application
Skills: Computer vision | Image Processing | Tensorflow | FastAPI | Streamlit | Python
Microscope imaging is vital to biological research. Enormous amount of images are being produced everyday. Precise delineation of cells in an image is essential for downstream analysis, like cell phenotype comparison and feature quantification. Given the overwhelming data volume, a high throughput image processing pipeline that takes in raw images and spits out a cell boundary binary file will be helpful for biological researchers. Another challenge for cell segmentation is that the discernibility for cells in phase contrast and differential interference contrast (DIC), the most used microscope imaging method, is poor to image processing software like Fiji ImageJ. Thresholding is only effective in detecting objects of high signal-to-noise ratio, like fluorescence signals, but not in DIC. The image processing pipeline should be able to recognize cells or target patterns out of low contrast images accurately.Machine learning (ML)-powered computer vision has became popular in both academy and industry. Most ML models are built based on convolutional neural networks (CNN) . I developed Cell Segger, an application having a tensorflow image processing flow and a pre-trained UNet model integrated. In the current version, only images in grayscale or color mode are accepted for an accurate prediction.
1. Cell Segger API
2. Cell Segger UI
